5.8 EMP_rrarefy
When performing second-generation high-throughput sequencing, the sequencing depth of different samples varies. Especially when analyzing microbiome data, the minimum sequencing depth of samples in the data is usually taken as the standard, and the data of all samples are resampled with replacement, to ensure that the sequencing depth of all samples is at one level.
5.8.1 Display current genomic sequencing depth
🏷️Example:
MAE |>
EMP_rrarefy(experiment = 'taxonomy',only_show_depth=T)

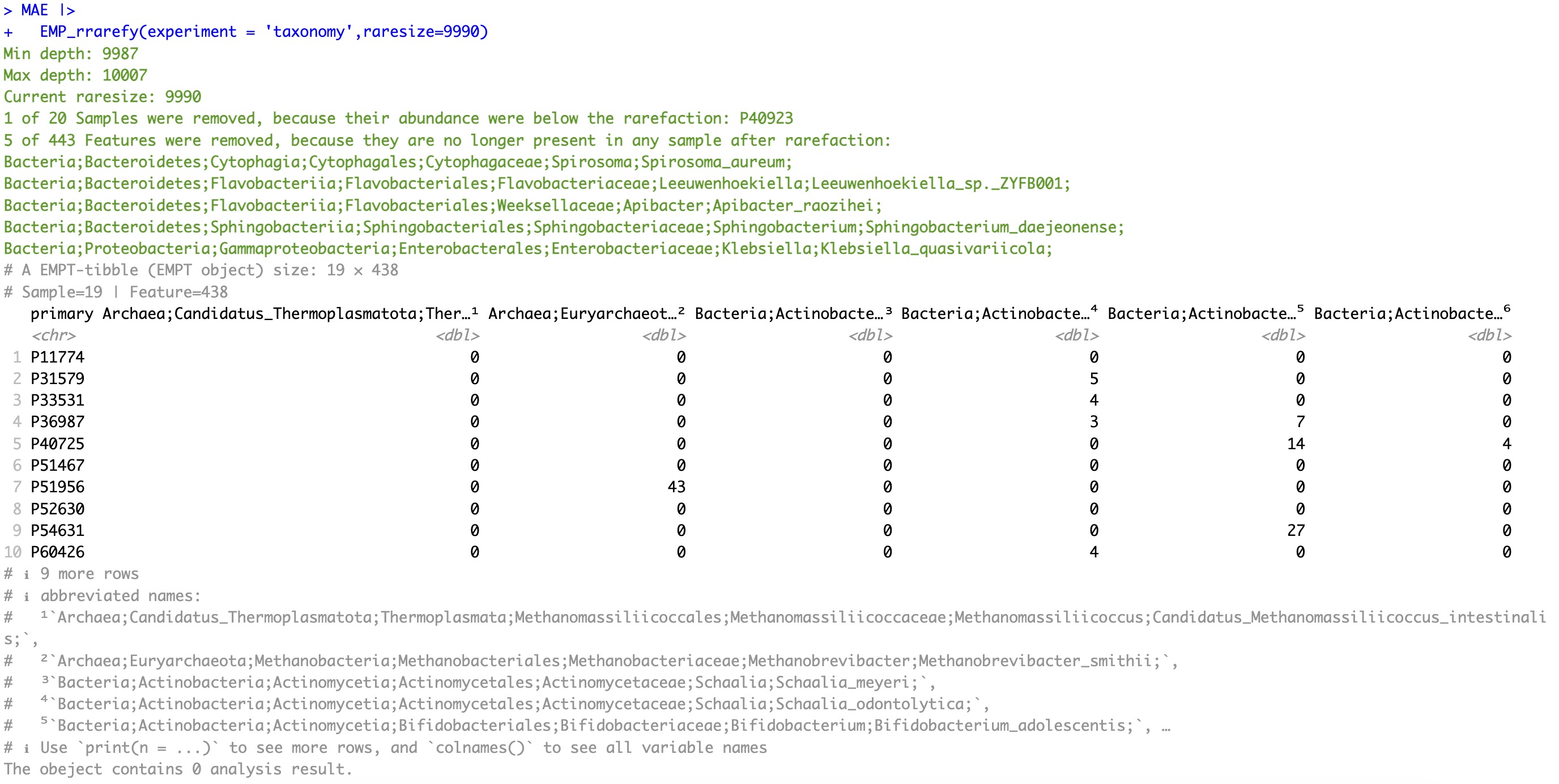
5.8.2 Resample at the minimum sequencing depth of samples within the current data
Note:
Since all samples reach the minimum sequencing depth threshold, the module
Since all samples reach the minimum sequencing depth threshold, the module
EMP_rrarefy does not filter any samples.
🏷️Example:
MAE |>
EMP_rrarefy(experiment = 'taxonomy')
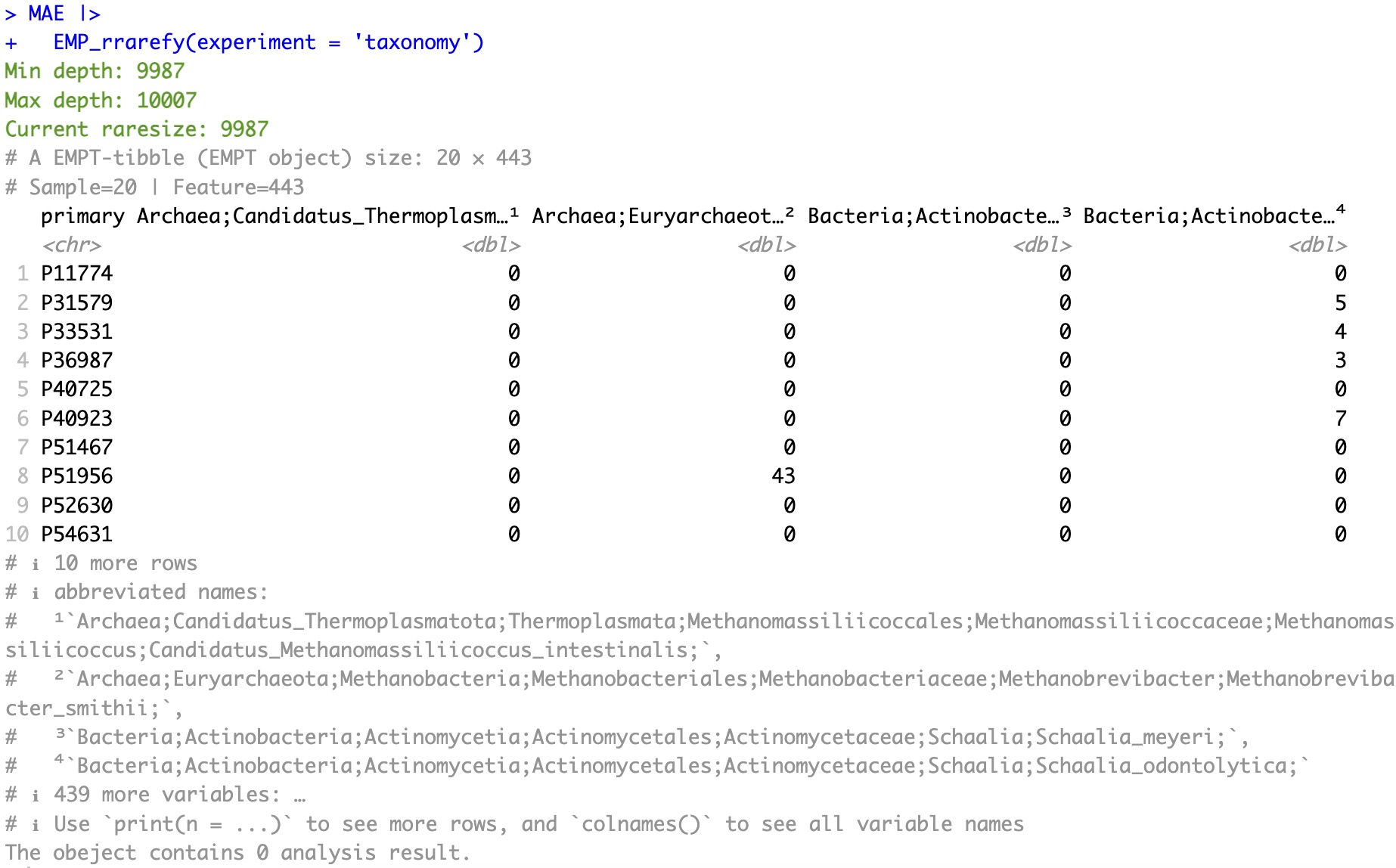
5.8.3 Resample according to specified sequencing depth
Note:
Users can customize the sequencing depth threshold by specifying the parameter
Users can customize the sequencing depth threshold by specifying the parameter
rasize, and any samples below this sequencing depth threshold will be filtered. At the same time, if some features are not present in all samples after resampling (i.e., the data is 0), these features will also be filtered.
MAE |>
EMP_rrarefy(experiment = 'taxonomy',raresize=9990)